Note
Click here to download the full example code
Fit Multiple Data Sets¶
Fitting multiple (simulated) Gaussian data sets simultaneously.
All minimizers require the residual array to be one-dimensional. Therefore, in
the objective we need to `flatten` the array before returning it.
TODO: this should be using the Model interface / built-in models!
import matplotlib.pyplot as plt
import numpy as np
from lmfit import Parameters, minimize, report_fit
def gauss(x, amp, cen, sigma):
"""Gaussian lineshape."""
return amp * np.exp(-(x-cen)**2 / (2.*sigma**2))
def gauss_dataset(params, i, x):
"""Calculate Gaussian lineshape from parameters for data set."""
amp = params['amp_%i' % (i+1)]
cen = params['cen_%i' % (i+1)]
sig = params['sig_%i' % (i+1)]
return gauss(x, amp, cen, sig)
def objective(params, x, data):
"""Calculate total residual for fits of Gaussians to several data sets."""
ndata, _ = data.shape
resid = 0.0*data[:]
# make residual per data set
for i in range(ndata):
resid[i, :] = data[i, :] - gauss_dataset(params, i, x)
# now flatten this to a 1D array, as minimize() needs
return resid.flatten()
Create five simulated Gaussian data sets
x = np.linspace(-1, 2, 151)
data = []
for i in np.arange(5):
params = Parameters()
amp = 0.60 + 9.50*np.random.rand()
cen = -0.20 + 1.20*np.random.rand()
sig = 0.25 + 0.03*np.random.rand()
dat = gauss(x, amp, cen, sig) + np.random.normal(size=x.size, scale=0.1)
data.append(dat)
data = np.array(data)
Create five sets of fitting parameters, one per data set
Constrain the values of sigma to be the same for all peaks by assigning sig_2, …, sig_5 to be equal to sig_1.
for iy in (2, 3, 4, 5):
fit_params['sig_%i' % iy].expr = 'sig_1'
Run the global fit and show the fitting result
Out:
[[Variables]]
amp_1: 6.35769745 +/- 0.02471391 (0.39%) (init = 0.5)
cen_1: -0.06087255 +/- 0.00141148 (2.32%) (init = 0.4)
sig_1: 0.27096733 +/- 6.7335e-04 (0.25%) (init = 0.3)
amp_2: 6.23505243 +/- 0.02466565 (0.40%) (init = 0.5)
cen_2: 0.90331537 +/- 0.00143924 (0.16%) (init = 0.4)
sig_2: 0.27096733 +/- 6.7335e-04 (0.25%) == 'sig_1'
amp_3: 6.74510330 +/- 0.02487197 (0.37%) (init = 0.5)
cen_3: 0.30698606 +/- 0.00133040 (0.43%) (init = 0.4)
sig_3: 0.27096733 +/- 6.7335e-04 (0.25%) == 'sig_1'
amp_4: 3.62962270 +/- 0.02384778 (0.66%) (init = 0.5)
cen_4: 0.00271542 +/- 0.00247235 (91.05%) (init = 0.4)
sig_4: 0.27096733 +/- 6.7335e-04 (0.25%) == 'sig_1'
amp_5: 6.29975266 +/- 0.02469084 (0.39%) (init = 0.5)
cen_5: -0.15885172 +/- 0.00142452 (0.90%) (init = 0.4)
sig_5: 0.27096733 +/- 6.7335e-04 (0.25%) == 'sig_1'
[[Correlations]] (unreported correlations are < 0.100)
C(sig_1, amp_3) = -0.337
C(amp_1, sig_1) = -0.320
C(sig_1, amp_5) = -0.317
C(sig_1, amp_2) = -0.314
C(sig_1, amp_4) = -0.189
C(amp_1, amp_3) = 0.108
C(amp_3, amp_5) = 0.107
C(amp_2, amp_3) = 0.106
C(amp_1, amp_5) = 0.101
C(amp_1, amp_2) = 0.100
Plot the data sets and fits
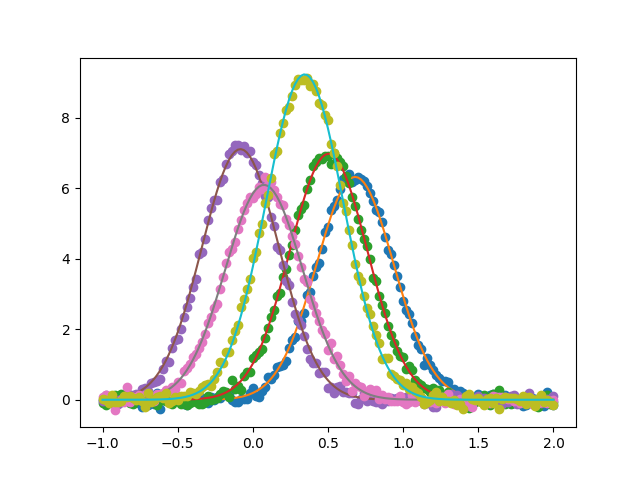
Out:
/Users/Newville/Codes/lmfit-py/examples/example_fit_multi_datasets.py:88: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
Total running time of the script: ( 0 minutes 0.139 seconds)