Note
Click here to download the full example code
Fit with Algebraic Constraint¶
Out:
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 53
# data points = 601
# variables = 6
chi-square = 71878.3055
reduced chi-square = 120.803875
Akaike info crit = 2887.26503
Bayesian info crit = 2913.65660
[[Variables]]
amp_g: 21.1877704 +/- 0.32191824 (1.52%) (init = 10)
cen_g: 8.11125923 +/- 0.01162984 (0.14%) (init = 9)
wid_g: 1.20925846 +/- 0.01170853 (0.97%) (init = 1)
amp_tot: 30.6003738 +/- 0.36481391 (1.19%) (init = 20)
amp_l: 9.41260340 +/- 0.61672681 (6.55%) == 'amp_tot - amp_g'
cen_l: 9.61125923 +/- 0.01162984 (0.12%) == '1.5+cen_g'
wid_l: 2.41851692 +/- 0.02341706 (0.97%) == '2*wid_g'
line_slope: 0.49615727 +/- 0.00170178 (0.34%) (init = 0)
line_off: 0.04128591 +/- 0.02448064 (59.30%) (init = 0)
[[Correlations]] (unreported correlations are < 0.100)
C(amp_g, wid_g) = 0.866
C(amp_g, cen_g) = 0.750
C(line_slope, line_off) = -0.714
C(cen_g, amp_tot) = -0.695
C(cen_g, wid_g) = 0.623
C(amp_g, amp_tot) = -0.612
C(amp_tot, line_off) = -0.588
C(wid_g, amp_tot) = -0.412
C(cen_g, line_off) = 0.387
C(amp_g, line_off) = 0.183
C(amp_g, line_slope) = 0.183
C(wid_g, line_slope) = 0.174
/Users/Newville/Codes/lmfit-py/examples/example_fit_with_algebraic_constraint.py:66: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
import matplotlib.pyplot as plt
from numpy import linspace, random
from lmfit import Minimizer, Parameters
from lmfit.lineshapes import gaussian, lorentzian
from lmfit.printfuncs import report_fit
def residual(pars, x, sigma=None, data=None):
yg = gaussian(x, pars['amp_g'], pars['cen_g'], pars['wid_g'])
yl = lorentzian(x, pars['amp_l'], pars['cen_l'], pars['wid_l'])
slope = pars['line_slope']
offset = pars['line_off']
model = yg + yl + offset + x*slope
if data is None:
return model
if sigma is None:
return model - data
return (model - data) / sigma
random.seed(0)
x = linspace(0.0, 20.0, 601)
data = (gaussian(x, 21, 8.1, 1.2) +
lorentzian(x, 10, 9.6, 2.4) +
random.normal(scale=0.23, size=x.size) +
x*0.5)
pfit = Parameters()
pfit.add(name='amp_g', value=10)
pfit.add(name='cen_g', value=9)
pfit.add(name='wid_g', value=1)
pfit.add(name='amp_tot', value=20)
pfit.add(name='amp_l', expr='amp_tot - amp_g')
pfit.add(name='cen_l', expr='1.5+cen_g')
pfit.add(name='wid_l', expr='2*wid_g')
pfit.add(name='line_slope', value=0.0)
pfit.add(name='line_off', value=0.0)
sigma = 0.021 # estimate of data error (for all data points)
myfit = Minimizer(residual, pfit,
fcn_args=(x,), fcn_kws={'sigma': sigma, 'data': data},
scale_covar=True)
result = myfit.leastsq()
init = residual(pfit, x)
fit = residual(result.params, x)
report_fit(result)
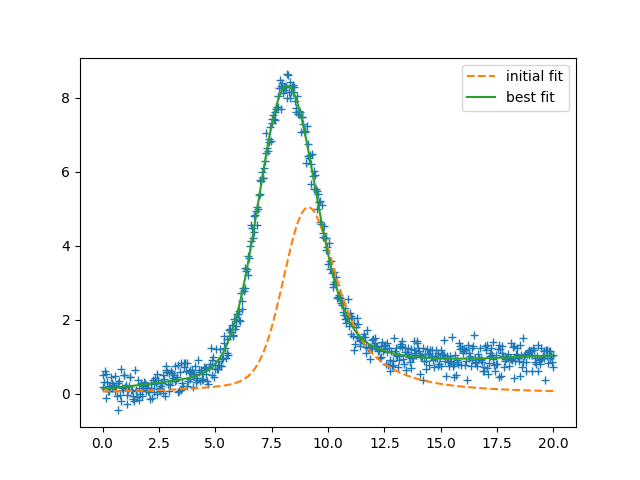
plt.plot(x, data, 'r+')
plt.plot(x, init, 'b--', label='initial fit')
plt.plot(x, fit, 'k-', label='best fit')
plt.legend(loc='best')
plt.show()
Total running time of the script: ( 0 minutes 0.103 seconds)