Note
Click here to download the full example code
Fit Using Bounds¶
A major advantage of using lmfit is that one can specify boundaries on fitting parameters, even if the underlying algorithm in SciPy does not support this. For more information on how this is implemented, please refer to: https://lmfit.github.io/lmfit-py/bounds.html
The example below shows how to set boundaries using the min and max
attributes to fitting parameters.
import matplotlib.pyplot as plt
from numpy import exp, linspace, pi, random, sign, sin
from lmfit import Parameters, minimize
from lmfit.printfuncs import report_fit
p_true = Parameters()
p_true.add('amp', value=14.0)
p_true.add('period', value=5.4321)
p_true.add('shift', value=0.12345)
p_true.add('decay', value=0.01000)
def residual(pars, x, data=None):
argu = (x * pars['decay'])**2
shift = pars['shift']
if abs(shift) > pi/2:
shift = shift - sign(shift)*pi
model = pars['amp'] * sin(shift + x/pars['period']) * exp(-argu)
if data is None:
return model
return model - data
random.seed(0)
x = linspace(0, 250, 1500)
noise = random.normal(scale=2.80, size=x.size)
data = residual(p_true, x) + noise
fit_params = Parameters()
fit_params.add('amp', value=13.0, max=20, min=0.0)
fit_params.add('period', value=2, max=10)
fit_params.add('shift', value=0.0, max=pi/2., min=-pi/2.)
fit_params.add('decay', value=0.02, max=0.10, min=0.00)
out = minimize(residual, fit_params, args=(x,), kws={'data': data})
fit = residual(out.params, x)
This gives the following fitting results:
report_fit(out, show_correl=True, modelpars=p_true)
Out:
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 74
# data points = 1500
# variables = 4
chi-square = 11301.3646
reduced chi-square = 7.55438813
Akaike info crit = 3037.18756
Bayesian info crit = 3058.44044
[[Variables]]
amp: 13.8903938 +/- 0.24412383 (1.76%) (init = 13), model_value = 14
period: 5.44026442 +/- 0.01416175 (0.26%) (init = 2), model_value = 5.4321
shift: 0.12464470 +/- 0.02414209 (19.37%) (init = 0), model_value = 0.12345
decay: 0.00996351 +/- 2.0278e-04 (2.04%) (init = 0.02), model_value = 0.01
[[Correlations]] (unreported correlations are < 0.100)
C(period, shift) = 0.800
C(amp, decay) = 0.576
and shows the plot below:
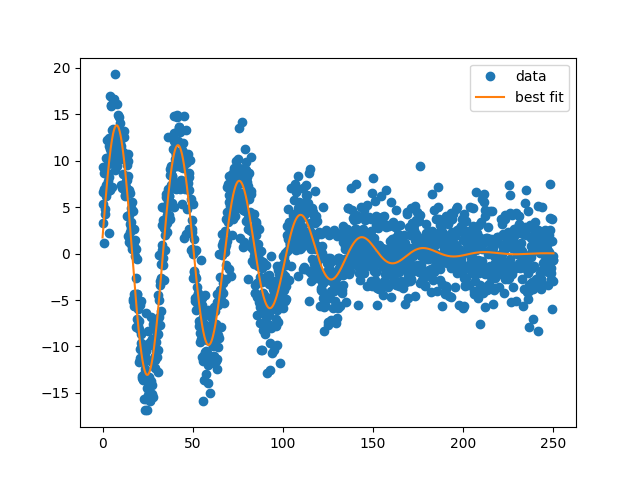
Out:
/Users/Newville/Codes/lmfit-py/examples/example_fit_with_bounds.py:62: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
Total running time of the script: ( 0 minutes 0.084 seconds)