Note
Click here to download the full example code
doc_model_uncertainty.py¶
Out:
[[Model]]
Model(gaussian)
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 33
# data points = 101
# variables = 3
chi-square = 3.40883599
reduced chi-square = 0.03478404
Akaike info crit = -336.263713
Bayesian info crit = -328.418352
[[Variables]]
amp: 8.88021830 +/- 0.11359492 (1.28%) (init = 5)
cen: 5.65866102 +/- 0.01030495 (0.18%) (init = 5)
wid: 0.69765468 +/- 0.01030495 (1.48%) (init = 1)
[[Correlations]] (unreported correlations are < 0.100)
C(amp, wid) = 0.577
##
import warnings
warnings.filterwarnings("ignore")
##
# <examples/doc_model_uncertainty.py>
import matplotlib.pyplot as plt
from numpy import exp, loadtxt, pi, sqrt
from lmfit import Model
data = loadtxt('model1d_gauss.dat')
x = data[:, 0]
y = data[:, 1]
def gaussian(x, amp, cen, wid):
"""1-d gaussian: gaussian(x, amp, cen, wid)"""
return (amp / (sqrt(2*pi) * wid)) * exp(-(x-cen)**2 / (2*wid**2))
gmodel = Model(gaussian)
result = gmodel.fit(y, x=x, amp=5, cen=5, wid=1)
print(result.fit_report())
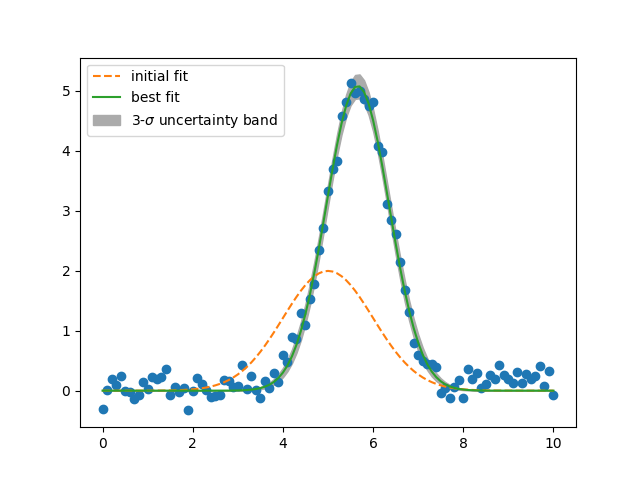
dely = result.eval_uncertainty(sigma=3)
plt.plot(x, y, 'bo')
plt.plot(x, result.init_fit, 'k--', label='initial fit')
plt.plot(x, result.best_fit, 'r-', label='best fit')
plt.fill_between(x, result.best_fit-dely, result.best_fit+dely,
color="#ABABAB", label=r'3-$\sigma$ uncertainty band')
plt.legend(loc='best')
plt.show()
# <end examples/doc_model_uncertainty.py>
Total running time of the script: ( 0 minutes 0.117 seconds)