2.4 MCA Displays
There are three functions under this group.†
2.4.1
Preferences
This window allows you to change the display preferences, such as changing the vertical scale, horizontal scale, etc., without affecting the data file.
2.4.2 JCPDS
This allows you to select the JCPDS diffraction file for a given material.† Note that although many materials are already in the collection of the JCPDS files, your sample file may not exist.† To create a new JCPDS file, following this link.
2.4.3
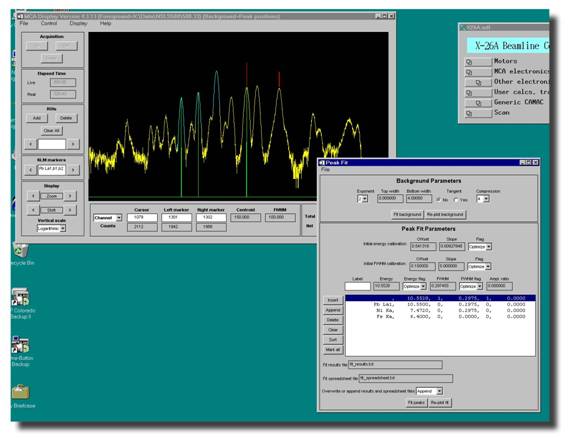
Peak fitting using the new MCA routine is also significantly
easier than it once was. First thing you need to do is call up the file you
want to fit. Then open up the peak fitting routines by selecting Display from the menu, and then
In the next section you'll tell the
MCA program what peaks to fit. Unlike the ROIs, here
you have no limit on the number of peaks you can define. The first peak that
will be listed will have no Label and the energy will be meaningless, like the
line highlighted in blue in the example above. This is normal, what it's saying is that...OK here's a peak waiting to be defined
by you! So let's define it. Highlight the peak (it'll be marked in blue), and
in the Label box above enter the name of the
peak you wish to fit. Again, follow our naming conventions (i.e. Fe Ka for iron k-alpha, Mn Kb
for manganese k-beta, Pb La1 for lead L-alpha 1 etc.) After you've entered
the name hit the Enter key and what you'll
see is that the name of the line selected in blue will change to reflect this
and that the information about this peak (energy, FWHM, etc.) will change to
correspond to the values of any peak that the routine found at this position in
your spectra. You'll also see that in your spectral display the peak will now
be marked with a single green vertical line. If you would rather enter energies
instead of the names of the fluorescence lines, just enter the number in the Label field and hit Enter.
To add another peak now click on the Insert button. Now select this line (it'll be
marked in blue) and 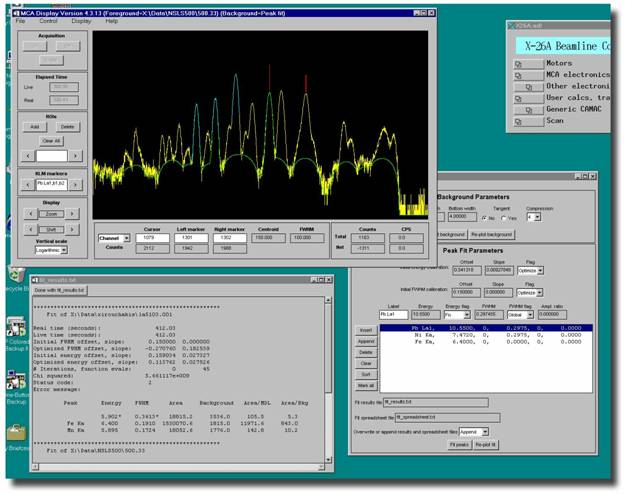 enter its information. Repeat this
procedure until all your peaks are defined. To change a peak simply select it
(it'll be marked in blue) and change its Label. The buttons to the left allow
you to manipulate the defined peak positions and should be self-explanatory.
Once you've defined all your peaks you have the option of saving this list or
recalling a previously saved list of peaks by selecting File and then either
enter its information. Repeat this
procedure until all your peaks are defined. To change a peak simply select it
(it'll be marked in blue) and change its Label. The buttons to the left allow
you to manipulate the defined peak positions and should be self-explanatory.
Once you've defined all your peaks you have the option of saving this list or
recalling a previously saved list of peaks by selecting File and then either
Now we're ready to fit the
peaks.† Click on that
†